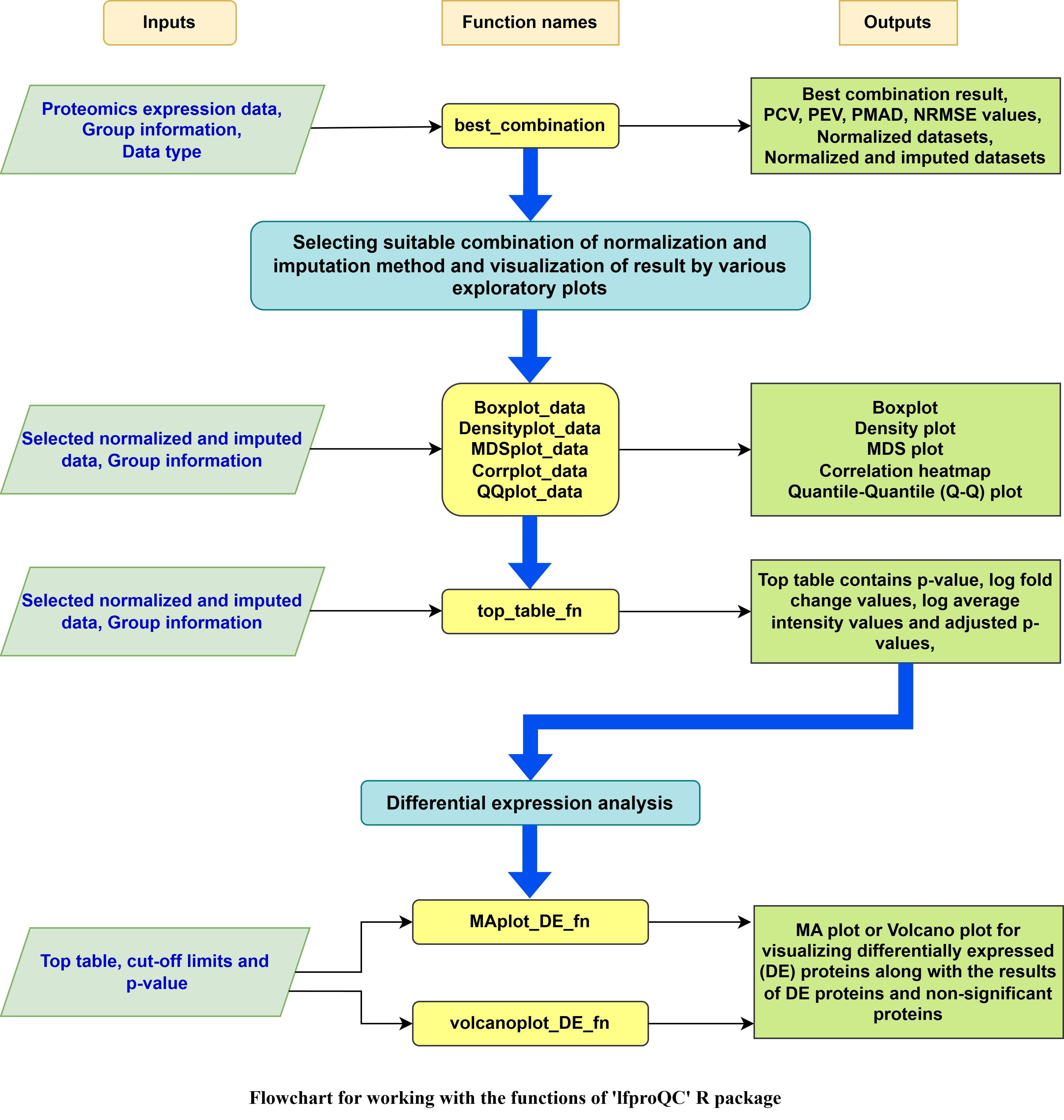
The goal of lfproQC R package is to provide an optimal combination of normalization and imputation methods for the label-free proteomics expression dataset.
You can install the development version of lfproQC from GitHub with:
# install.packages("devtools")
devtools::install_github("kabilansbio/lfproQC", build_manual = TRUE, build_vignette = TRUE)This is a basic example for finding the best combinations of normalization and imputation method for the label-free proteomics expression dataset:
library(lfproQC)
## basic example code with the example dataset and data groups
yeast <- best_combination(yeast_data, yeast_groups, data_type = "Protein")
yeast$`Best combinations`
#> PCV_best_combination PEV_best_combination PMAD_best_combination
#> 1 rlr_knn, rlr_lls vsn_lls rlr_llsThe overall workflow for using the ‘lfproQC’ package
