

The package contains three functions to crunch SHAP values:
permshap(): Permutation SHAP algorithm
of [1]. Both exact and sampling versions are available.kernelshap(): Kernel SHAP algorithm of
[2] and [3]. Both exact and (partly exact) sampling versions are
available.additive_shap(): For additive
models fitted via lm(), glm(),
mgcv::gam(), mgcv::bam(),
gam::gam(), survival::coxph(), or
survival::survreg(). Exponentially faster than the
model-agnostic options above, and recommended if possible.To explain your model, select an explanation dataset X
(up to 1000 rows from the training data, feature columns only). Use
{shapviz} to visualize the resulting SHAP values.
Remarks to permshap() and
kernelshap()
bg_X to calculate marginal means (up to 500 rows from the
training data). In cases with a natural “off” value (like MNIST digits),
this can also be a single row with all values set to the off value. If
unspecified, 200 rows are randomly sampled from X.kernelshap() switches to a partly exact algorithm with
faster convergence than the sampling version of permutation SHAP.permshap() and
kernelshap() give the same results as
additive_shap as long as the full training data would be
used as background data.# From CRAN
install.packages("kernelshap")
# Or the development version:
devtools::install_github("ModelOriented/kernelshap")Let’s model diamond prices with a random forest. As an alternative, you could use the {treeshap} package in this situation.
library(kernelshap)
library(ggplot2)
library(ranger)
library(shapviz)
options(ranger.num.threads = 8)
diamonds <- transform(
diamonds,
log_price = log(price),
log_carat = log(carat)
)
xvars <- c("log_carat", "clarity", "color", "cut")
fit <- ranger(
log_price ~ log_carat + clarity + color + cut,
data = diamonds,
num.trees = 100,
seed = 20
)
fit # OOB R-squared 0.989
# 1) Sample rows to be explained
set.seed(10)
X <- diamonds[sample(nrow(diamonds), 1000), xvars]
# 2) Optional: Select background data. If unspecified, 200 rows from X are used
bg_X <- diamonds[sample(nrow(diamonds), 200), ]
# 3) Crunch SHAP values (22 seconds)
# Since the number of features is small, we use permshap()
system.time(
ps <- permshap(fit, X, bg_X = bg_X)
)
ps
# SHAP values of first observations:
log_carat clarity color cut
[1,] 1.1913247 0.09005467 -0.13430720 0.000682593
[2,] -0.4931989 -0.11724773 0.09868921 0.028563613
# Indeed, Kernel SHAP gives the same:
ks <- kernelshap(fit, X, bg_X = bg_X)
ks
log_carat clarity color cut
[1,] 1.1913247 0.09005467 -0.13430720 0.000682593
[2,] -0.4931989 -0.11724773 0.09868921 0.028563613
# 4) Analyze with {shapviz}
ps <- shapviz(ps)
sv_importance(ps)
sv_dependence(ps, xvars)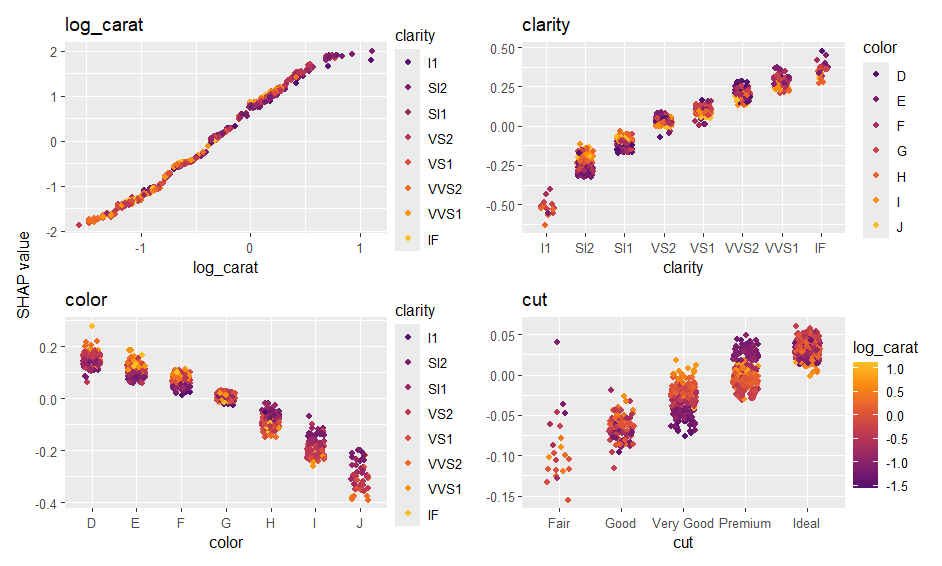
The {kernelshap} package can deal with almost any situation. We will show some of the flexibility here. The first two examples require you to run at least up to Step 2 of the “Basic Usage” code.
Parallel computing for permshap() and
kernelshap() is supported via {doFuture} and {foreach}.
Note that this does not work for all models.
On Windows, sometimes not all packages or global objects are passed
to the parallel sessions. Often, this can be fixed via
parallel_args using the arguments “packages” and “globals”
passed to foreach(.options.future = ...), see this
example:
library(doFuture)
library(mgcv)
plan(multisession, workers = 4) # Windows
# plan(multicore, workers = 4) # Linux, macOS, Solaris
# GAM with interactions - we cannot use additive_shap()
fit <- gam(log_price ~ s(log_carat) + clarity * color + cut, data = diamonds)
system.time( # 4 seconds in parallel
ps <- permshap(
fit, X, bg_X = bg_X, parallel = TRUE, parallel_args = list(packages = "mgcv")
)
)
ps
# SHAP values of first observations:
# log_carat clarity color cut
# [1,] 1.26801 0.1023518 -0.09223291 0.004512402
# [2,] -0.51546 -0.1174766 0.11122775 0.030243973
# Because there are no interactions of order above 2, Kernel SHAP gives the same:
system.time( # 12 s non-parallel
ks <- kernelshap(fit, X, bg_X = bg_X)
)
all.equal(ps$S, ks$S)
# [1] TRUE
# Now the usual plots:
sv <- shapviz(ps)
sv_importance(sv, kind = "bee")
sv_dependence(sv, xvars)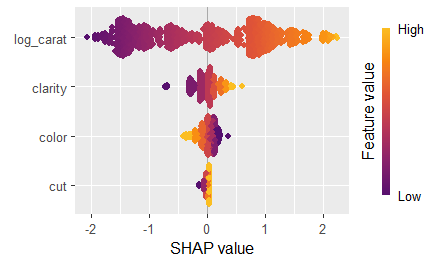
In this {keras} example, we show how to use a tailored
predict() function that complies with
(The results are not fully reproducible.)
library(keras)
nn <- keras_model_sequential()
nn |>
layer_dense(units = 30, activation = "relu", input_shape = 4) |>
layer_dense(units = 15, activation = "relu") |>
layer_dense(units = 1)
nn |>
compile(optimizer = optimizer_adam(0.001), loss = "mse")
cb <- list(
callback_early_stopping(patience = 20),
callback_reduce_lr_on_plateau(patience = 5)
)
nn |>
fit(
x = data.matrix(diamonds[xvars]),
y = diamonds$log_price,
epochs = 100,
batch_size = 400,
validation_split = 0.2,
callbacks = cb
)
pred_fun <- function(mod, X)
predict(mod, data.matrix(X), batch_size = 1e4, verbose = FALSE, workers = 4)
system.time( # 42 s
ps <- permshap(nn, X, bg_X = bg_X, pred_fun = pred_fun)
)
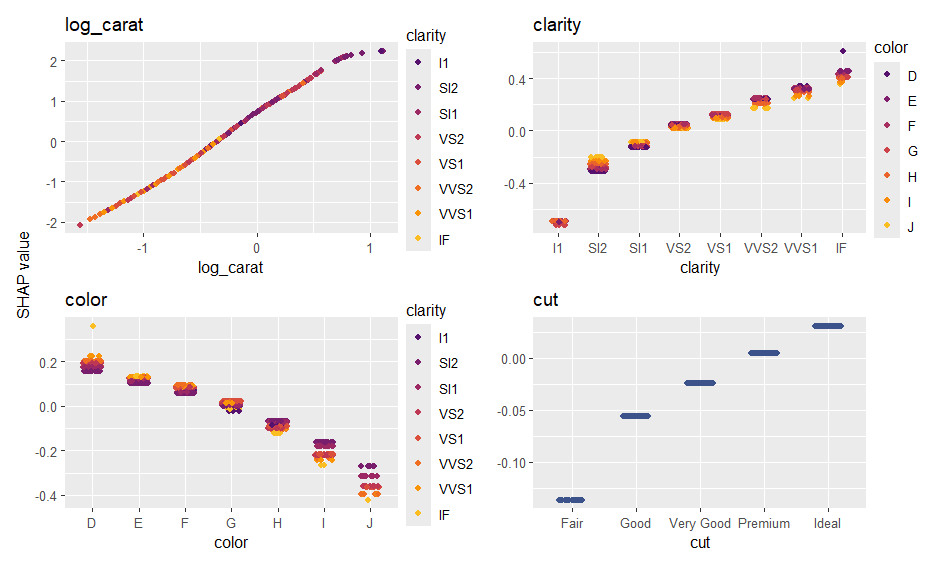
ps <- shapviz(ps)
sv_importance(ps, show_numbers = TRUE)
sv_dependence(ps, xvars)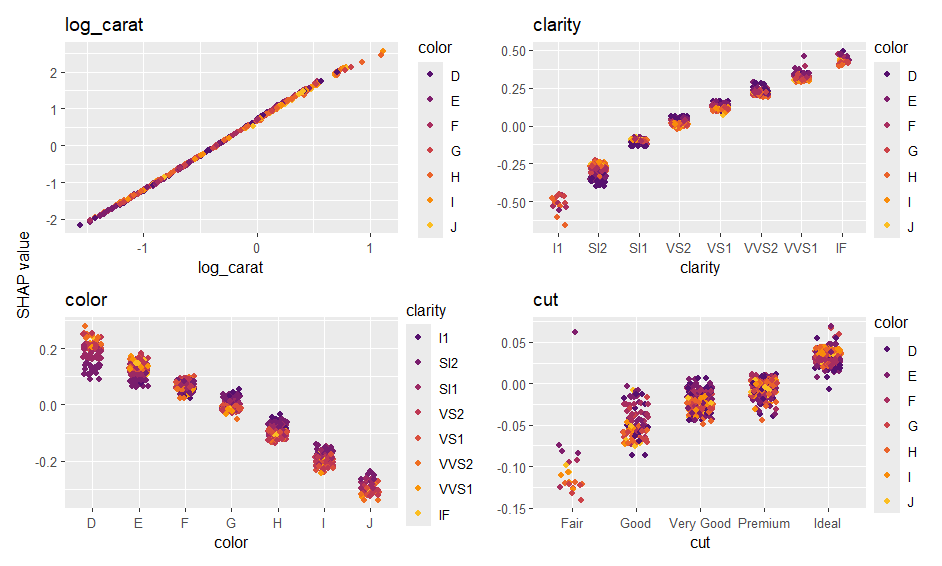
The additive explainer extracts the additive contribution of each feature from a model of suitable class.
fit <- lm(log(price) ~ log(carat) + color + clarity + cut, data = diamonds)
shap_values <- additive_shap(fit, diamonds) |>
shapviz()
sv_importance(shap_values)
sv_dependence(shap_values, v = "carat", color_var = NULL){kernelshap} supports multivariate predictions like:
Here, we use the iris data (no need to run code from
above).
library(kernelshap)
library(ranger)
library(shapviz)
set.seed(1)
# Probabilistic classification
fit_prob <- ranger(Species ~ ., data = iris, probability = TRUE)
ps_prob <- permshap(fit_prob, X = iris[-5]) |>
shapviz()
sv_importance(ps_prob)
sv_dependence(ps_prob, "Petal.Length")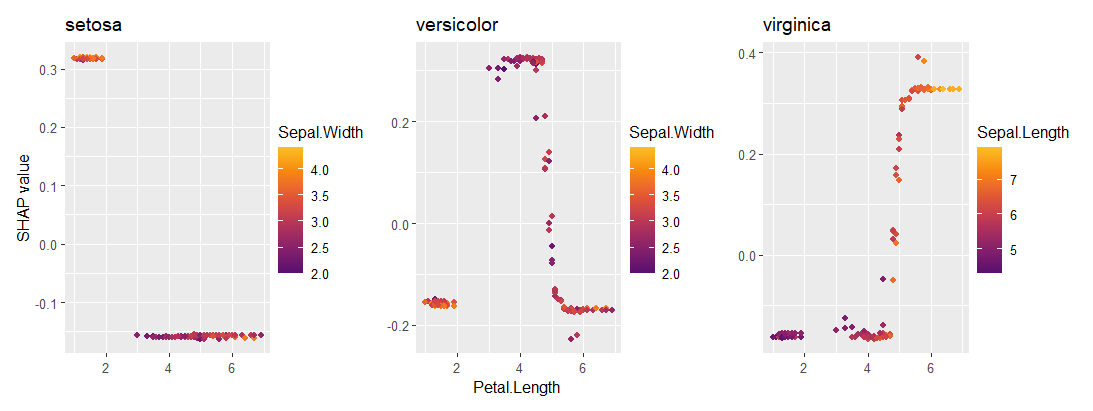
Meta-learning packages like {tidymodels}, {caret} or {mlr3} are
straightforward to use. The following examples additionally shows that
the ... arguments of permshap() and
kernelshap() are passed to predict().
library(kernelshap)
library(tidymodels)
set.seed(1)
iris_recipe <- iris |>
recipe(Species ~ .)
mod <- rand_forest(trees = 100) |>
set_engine("ranger") |>
set_mode("classification")
iris_wf <- workflow() |>
add_recipe(iris_recipe) |>
add_model(mod)
fit <- iris_wf |>
fit(iris)
system.time( # 3s
ps <- permshap(fit, iris[-5], type = "prob")
)
ps
# Some values
$.pred_setosa
Sepal.Length Sepal.Width Petal.Length Petal.Width
[1,] 0.02186111 0.012137778 0.3658278 0.2667667
[2,] 0.02628333 0.001315556 0.3683833 0.2706111library(kernelshap)
library(caret)
fit <- train(
Sepal.Length ~ .,
data = iris,
method = "lm",
tuneGrid = data.frame(intercept = TRUE),
trControl = trainControl(method = "none")
)
ps <- permshap(fit, iris[-1])library(kernelshap)
library(mlr3)
library(mlr3learners)
set.seed(1)
task_classif <- TaskClassif$new(id = "1", backend = iris, target = "Species")
learner_classif <- lrn("classif.rpart", predict_type = "prob")
learner_classif$train(task_classif)
x <- learner_classif$selected_features()
# Don't forget to pass predict_type = "prob" to mlr3's predict()
ps <- permshap(
learner_classif, X = iris, feature_names = x, predict_type = "prob"
)
ps
# $setosa
# Petal.Length Petal.Width
# [1,] 0.6666667 0
# [2,] 0.6666667 0[1] Erik Štrumbelj and Igor Kononenko. Explaining prediction models and individual predictions with feature contributions. Knowledge and Information Systems 41, 2014.
[2] Scott M. Lundberg and Su-In Lee. A Unified Approach to Interpreting Model Predictions. Advances in Neural Information Processing Systems 30, 2017.
[3] Ian Covert and Su-In Lee. Improving KernelSHAP: Practical Shapley Value Estimation Using Linear Regression. Proceedings of The 24th International Conference on Artificial Intelligence and Statistics, PMLR 130:3457-3465, 2021.