The fastcpd (fast change point detection) is a fast implmentation of change point detection methods in R. The fastcpd package is designed to find change points in a fast manner. It is easy to install and extensible to all kinds of change point problems with a user specified cost function apart from the built-in cost functions.
To learn more behind the algorithms:
install.packages(
"fastcpd",
repos = c("https://doccstat.r-universe.dev", "https://cloud.r-project.org")
)pak::pak("doccstat/fastcpd")
devtools::install_github("doccstat/fastcpd")# conda-forge is a fork from CRAN and may not be up-to-date
# Use mamba
mamba install r-fastcpd
# Use conda
conda install -c conda-forge r-fastcpdset.seed(1)
n <- 1000
x <- rep(0, n + 3)
for (i in 1:600) {
x[i + 3] <- 0.6 * x[i + 2] - 0.2 * x[i + 1] + 0.1 * x[i] + rnorm(1, 0, 3)
}
for (i in 601:1000) {
x[i + 3] <- 0.3 * x[i + 2] + 0.4 * x[i + 1] + 0.2 * x[i] + rnorm(1, 0, 3)
}
result <- fastcpd::fastcpd.ar(x[3 + seq_len(n)], 3, r.progress = FALSE)
summary(result)
#>
#> Call:
#> fastcpd::fastcpd.ar(data = x[3 + seq_len(n)], order = 3, r.progress = FALSE)
#>
#> Change points:
#> 614
#>
#> Cost values:
#> 2754.116 2038.945
#>
#> Parameters:
#> segment 1 segment 2
#> 1 0.57120256 0.2371809
#> 2 -0.20985108 0.4031244
#> 3 0.08221978 0.2290323
plot(result)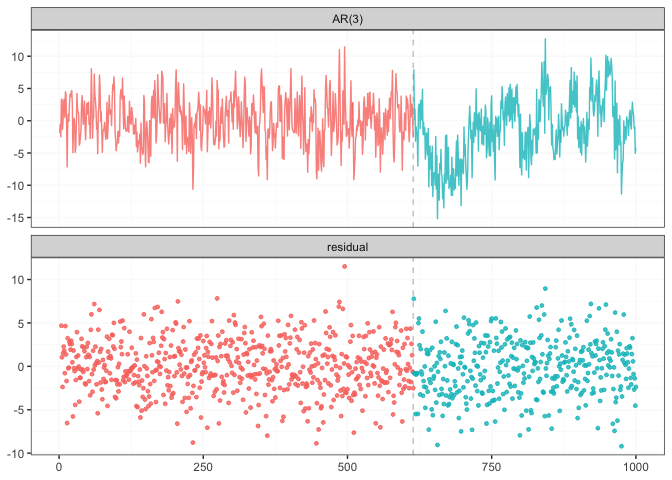
r.progress = FALSE is used to suppress the progress bar.
Users are expected to see the progress bar when running the code by
default.
library(microbenchmark)
set.seed(1)
n <- 5 * 10^6
mean_data <- c(rnorm(n / 2, 0, 1), rnorm(n / 2, 50, 1))
ggplot2::autoplot(microbenchmark(
fastcpd = fastcpd::fastcpd.mean(mean_data, r.progress = FALSE, cp_only = TRUE, variance_estimation = 1),
changepoint = changepoint::cpt.mean(mean_data, method = "PELT"),
fpop = fpop::Fpop(mean_data, 2 * log(n)),
gfpop = gfpop::gfpop(
data = mean_data,
mygraph = gfpop::graph(
penalty = 2 * log(length(mean_data)) * gfpop::sdDiff(mean_data) ^ 2,
type = "updown"
),
type = "mean"
),
jointseg = jointseg::jointSeg(mean_data, K = 12),
mosum = mosum::mosum(c(mean_data), G = 40),
not = not::not(mean_data, contrast = "pcwsConstMean"),
wbs = wbs::wbs(mean_data)
))
#> Warning in microbenchmark(fastcpd = fastcpd::fastcpd.mean(mean_data, r.progress
#> = FALSE, : less accurate nanosecond times to avoid potential integer overflows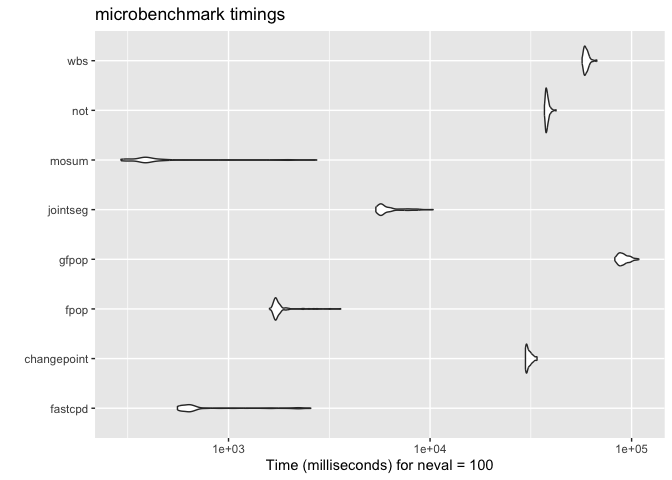
library(microbenchmark)
set.seed(1)
n <- 10^8
mean_data <- c(rnorm(n / 2, 0, 1), rnorm(n / 2, 50, 1))
system.time(fastcpd::fastcpd.mean(mean_data, r.progress = FALSE, cp_only = TRUE, variance_estimation = 1))
#> user system elapsed
#> 11.753 9.150 26.455
system.time(changepoint::cpt.mean(mean_data, method = "PELT"))
#> user system elapsed
#> 32.342 9.681 66.056
system.time(fpop::Fpop(mean_data, 2 * log(n)))
#> user system elapsed
#> 35.926 5.231 58.269
system.time(mosum::mosum(c(mean_data), G = 40))
#> user system elapsed
#> 5.518 11.516 38.368
ggplot2::autoplot(microbenchmark(
fastcpd = fastcpd::fastcpd.mean(mean_data, r.progress = FALSE, cp_only = TRUE, variance_estimation = 1),
changepoint = changepoint::cpt.mean(mean_data, method = "PELT"),
fpop = fpop::Fpop(mean_data, 2 * log(n)),
mosum = mosum::mosum(c(mean_data), G = 40),
times = 10
))
#> Warning in microbenchmark(fastcpd = fastcpd::fastcpd.mean(mean_data, r.progress
#> = FALSE, : less accurate nanosecond times to avoid potential integer overflows
Some packages are not included in the microbenchmark
comparison due to either memory constraints or long running time.
# Device: Mac mini (M1, 2020)
# Memory: 8 GB
system.time(CptNonPar::np.mojo(mean_data, G = floor(length(mean_data) / 6)))
#> Error: vector memory limit of 16.0 Gb reached, see mem.maxVSize()
#> Timing stopped at: 0.061 0.026 0.092
system.time(ecp::e.divisive(matrix(mean_data)))
#> Error: vector memory limit of 16.0 Gb reached, see mem.maxVSize()
#> Timing stopped at: 0.076 0.044 0.241
system.time(strucchange::breakpoints(y ~ 1, data = data.frame(y = mean_data)))
#> Timing stopped at: 265.1 145.8 832.5
system.time(breakfast::breakfast(mean_data))
#> Timing stopped at: 45.9 89.21 562.3fastcpd_arfastcpd_arimafastcpd_armafastcpd_garchfastcpd_varfastcpd_tsfastcpd_meanfastcpd_variancefastcpd_meanvariancefastcpd_binomialfastcpd_lassofastcpd_lmfastcpd_poissonvariance_armavariance_lmvariance_meanvariance_medianbitcoinoccupancytranscriptomeuk_seatbeltswell_logAvailable soon: rshiny.fastcpd.xingchi.li
The suggested packages are not required for the main functionality of the package. They are only required for the vignettes. If you want to learn more about the package comparison and other vignettes, you could either check out vignettes on CRAN or pkgdown generated documentation.
The package should be able to install on Mac and any Linux
distribution without any problems if all the dependencies are installed.
However, if you encountered problems related to gfortran, it might be
because RcppArmadillo is not installed previously. Try Mac OSX stackoverflow
solution or Linux
stackover solution if you have trouble installing
RcppArmadillo.
Fork the repo.
Create a new branch from main branch.
Make changes and commit them.
src/fastcpd_class_cost.cc with proper example and tests in
vignettes/gallery.Rmd and
tests/testthat/test-gallery.R.src/fastcpd_constants.h.R/fastcpd_wrappers.R for the new family of models and move
the examples to the new wrapper function as roxygen examples._pkgdown.yml.Push the changes to your fork.
Create a pull request.
Make sure the pull request does not create new warnings or errors
in devtools::check().