

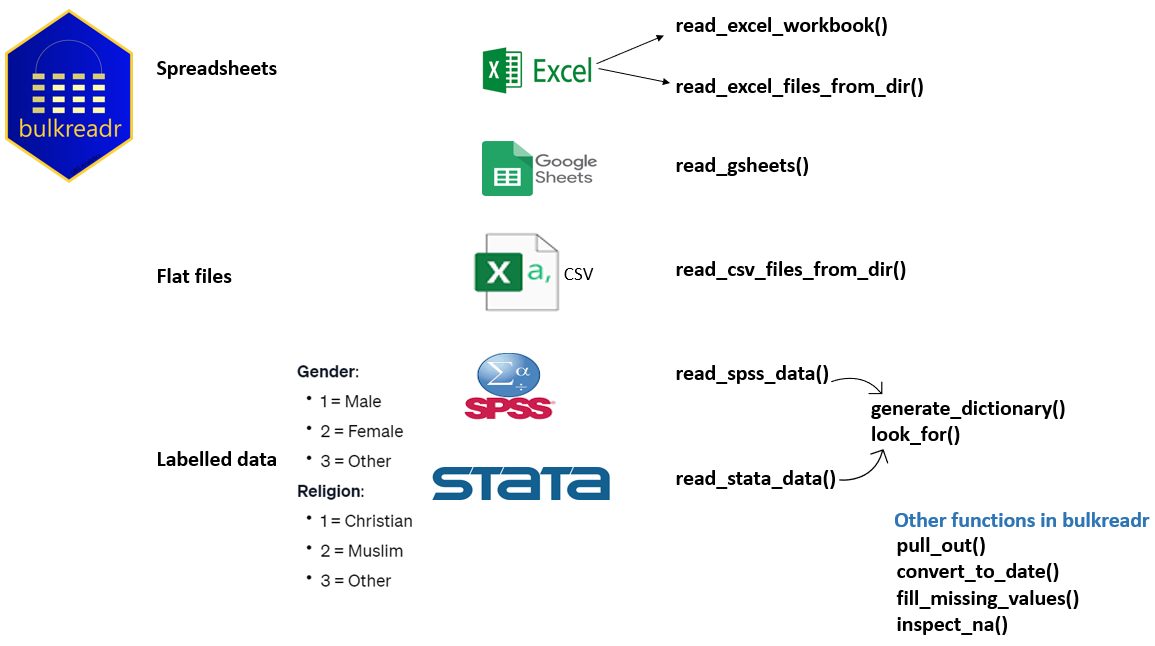
The Ultimate Tool for Reading Data in Bulk
bulkreadr is an R package designed to simplify and
streamline the process of reading and processing large volumes of data.
With a collection of functions tailored for bulk data operations, the
package allows users to efficiently read multiple sheets from Microsoft
Excel/Google Sheets workbooks and multiple CSV files from a directory.
It returns the data as organized data frames, making it convenient for
further analysis and manipulation.
Whether dealing with extensive data sets or batch processing tasks, “bulkreadr” empowers users to effortlessly handle data in bulk, saving time and effort in data preparation workflows.
Additionally, the package seamlessly works with labelled data from SPSS and Stata. For a quick video tutorial, I gave a talk at the International Association of Statistical Computing webinar. The recorded session is available here and the webinar resources here.
You can install bulkreadr package from CRAN with:
install.packages("bulkreadr")or the development version from GitHub with
if(!require("devtools")){
install.packages("devtools")
}
devtools::install_github("gbganalyst/bulkreadr")Now that you have installed bulkreadr package, you can
simply load it by using:
library(bulkreadr)To get started with bulkreadr, read the docs.
bulkreadr is designed to integrate with and augment the capabilities
of established packages such as readxl, readr,
and googlesheets4, offering enhanced functionality for
reading bulk data within the R programming environment.
readxl is the tidyverse package for reading Excel files (xls or xlsx) into an R data frame.
readr is the tidyverse package for reading delimited files (e.g., csv or tsv) into an R data frame.
googlesheets4 is the package to interact with Google Sheets through the Sheets API v4 https://developers.google.com/sheets/api.